10 Data preparation
Data preparation:
- Handle missing values
- Fix or remove outliers
- Feature selection
- Feature engineering
- Feature scaling
- Create a validation set
Next, we’ll preprocess our data before training the models. We mainly use the tidymodels packages recipes and workflows for this steps. Recipes are built as a series of optional data preparation steps, such as:
Data cleaning: Fix or remove outliers, fill in missing values (e.g., with zero, mean, median…) or drop their rows (or columns).
Feature selection: Drop the attributes that provide no useful information for the task.
Feature engineering: Discretize continuous features, decompose features (e.g., the weekday from a date variable, etc.), add promising transformations of features (e.g., log(x), sqrt(x), x2 , etc.) or aggregate features into promising new features (like we already did).
Feature scaling: Standardize or normalize features.
We will want to use our recipe across several steps as we train and test our models. To simplify this process, we can use a model workflow, which pairs a model and recipe together.
10.1 Data preparation
Before we create our recipes, we first select the variables which we will use in the model. Note that we keep longitude and latitude to be able to map the data in a later stage but we will not use the variables in our model.
housing_df_new <-
housing_df %>%
select( # select our predictors
longitude, latitude,
price_category,
median_income,
ocean_proximity,
bedrooms_per_room,
rooms_per_household,
population_per_household
)
glimpse(housing_df_new)#> Rows: 20,640
#> Columns: 8
#> $ longitude <dbl> -122, -122, -122, -122, -122, -122, -122, -1…
#> $ latitude <dbl> 38, 38, 38, 38, 38, 38, 38, 38, 38, 38, 38, …
#> $ price_category <fct> above, above, above, above, above, above, ab…
#> $ median_income <dbl> 8.3, 8.3, 7.3, 5.6, 3.8, 4.0, 3.7, 3.1, 2.1,…
#> $ ocean_proximity <fct> NEAR BAY, NEAR BAY, NEAR BAY, NEAR BAY, NEAR…
#> $ bedrooms_per_room <dbl> 0.15, 0.16, 0.13, 0.18, 0.17, 0.23, 0.19, 0.…
#> $ rooms_per_household <dbl> 7.0, 6.2, 8.3, 5.8, 6.3, 4.8, 4.9, 4.8, 4.3,…
#> $ population_per_household <dbl> 2.6, 2.1, 2.8, 2.5, 2.2, 2.1, 2.1, 1.8, 2.0,…Furthermore, we need to make a new data split since we updated the original data.
set.seed(123)
data_split <- initial_split(housing_df_new, # updated data
prop = 3/4,
strata = price_category)
train_data <- training(data_split)
test_data <- testing(data_split)10.2 Data prepropecessing recipe
The type of data preprocessing is dependent on the data and the type of model being fit. The excellent book “Tidy Modeling with R” provides an appendix with recommendations for baseline levels of preprocessing that are needed for various model functions (Kuhn and Silge (2021))
Let’s create a base recipe for all of our classification models. Note that the sequence of steps matter:
-
The
recipe()function has two arguments:- A formula. Any variable on the left-hand side of the tilde (
~) is considered the model outcome (here,price_category). On the right-hand side of the tilde are the predictors. Variables may be listed by name (separated by a+), or you can use the dot (.) to indicate all other variables as predictors. - The data. A recipe is associated with the data set used to create the model. This will typically be the training set, so
data = train_datahere.
- A formula. Any variable on the left-hand side of the tilde (
update_role(): This step of adding roles to a recipe is optional; the purpose of using it here is that those two variables can be retained in the data but not included in the model. This can be convenient when, after the model is fit, we want to investigate some poorly predicted value. These ID columns will be available and can be used to try to understand what went wrong.step_naomit()removes observations (rows of data) if they contain NA or NaN values. We useskip = TRUEbecause we don’t want to perform this part to new data so that the number of samples in the assessment set is the same as the number of predicted values (even if they are NA).
Note that instead of deleting missing values we could also easily substitute (i.e., impute) missing values of variables by one of the following methods (using the training set):
Take a look at the recipes reference for an overview about all possible imputation methods.
step_novel()converts all nominal variables to factors and takes care of other issues related to categorical variables.step_log()will log transform data (since some of our numerical variables are right-skewed). Note that this step can not be performed on negative numbers.step_normalize()normalizes (center and scales) the numeric variables to have a standard deviation of one and a mean of zero. (i.e., z-standardization).step_dummy()converts our factor columnocean_proximityinto numeric binary (0 and 1) variables.
Note that this step may cause problems if your categorical variable has too many levels - especially if some of the levels are very infrequent. In this case you should either drop the variable or pool infrequently occurring values into an “other” category with step_other. This steps has to be performed befor step_dummy.
step_zv(): removes any numeric variables that have zero variance.step_corr(): will remove predictor variables that have large correlations with other predictor variables.
Note that the package themis contains extra steps for the recipes package for dealing with imbalanced data. A classification data set with skewed class proportions is called imbalanced. Classes that make up a large proportion of the data set are called majority classes. Those that make up a smaller proportion are minority classes (see Google Developers for more details). Themis provides various methods for over-sampling (e.g. SMOTE) and under-sampling. However, we don’t have to use this methods since our data is not imbalanced.
housing_rec <-
recipe(price_category ~ .,
data = train_data) %>%
update_role(longitude, latitude,
new_role = "ID") %>%
step_log(
median_income,
bedrooms_per_room, rooms_per_household,
population_per_household
) %>%
step_naomit(everything(), skip = TRUE) %>%
step_novel(all_nominal(), -all_outcomes()) %>%
step_normalize(all_numeric(), -all_outcomes(),
-longitude, -latitude) %>%
step_dummy(all_nominal(), -all_outcomes()) %>%
step_zv(all_numeric(), -all_outcomes()) %>%
step_corr(all_predictors(), threshold = 0.7, method = "spearman") To view the current set of variables and roles, use the summary() function:
summary(housing_rec)#> # A tibble: 8 x 4
#> variable type role source
#> <chr> <chr> <chr> <chr>
#> 1 longitude numeric ID original
#> 2 latitude numeric ID original
#> 3 median_income numeric predictor original
#> 4 ocean_proximity nominal predictor original
#> 5 bedrooms_per_room numeric predictor original
#> 6 rooms_per_household numeric predictor original
#> # … with 2 more rowsIf we would like to check if all of our preprocessing steps from above actually worked, we can proceed as follows:
prepped_data <-
housing_rec %>% # use the recipe object
prep() %>% # perform the recipe on training data
juice() # extract only the preprocessed dataframe Take a look at the data structure:
glimpse(prepped_data)#> Rows: 15,317
#> Columns: 10
#> $ longitude <dbl> -122, -122, -122, -122, -122, -122, -122, …
#> $ latitude <dbl> 38, 38, 38, 38, 38, 38, 38, 38, 38, 38, 38…
#> $ median_income <dbl> 1.85, 1.56, 1.03, 0.32, 0.11, -0.22, -1.08…
#> $ rooms_per_household <dbl> 0.6768, 1.7192, 0.4206, -0.3141, -0.1852, …
#> $ population_per_household <dbl> -1.073, -0.042, -0.388, -1.022, -1.041, -1…
#> $ price_category <fct> above, above, above, above, above, above, …
#> $ ocean_proximity_INLAND <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
#> $ ocean_proximity_ISLAND <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
#> $ ocean_proximity_NEAR.BAY <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
#> $ ocean_proximity_NEAR.OCEAN <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …Visualize the numerical data:
prepped_data %>%
select(price_category,
median_income,
rooms_per_household,
population_per_household) %>%
ggscatmat(corMethod = "spearman",
alpha=0.2)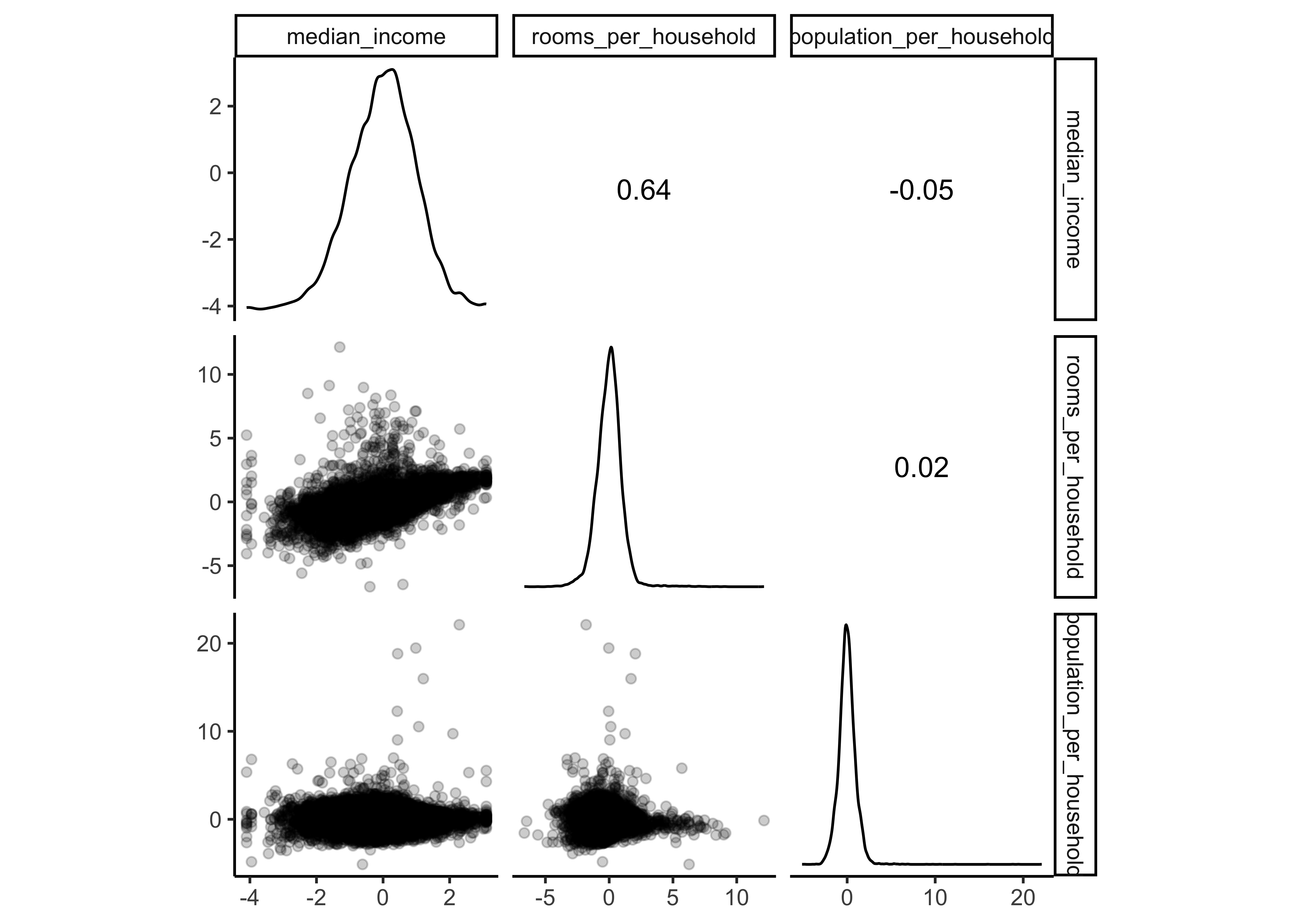
You should notice that:
the variables
longitudeandlatitudedid not change.median_income,rooms_per_householdandpopulation_per_householdare now z-standardized and the distributions are a bit less right skewed (due to our log transformation)ocean_proximitywas replaced by dummy variables.
10.3 Validation set
Remember that we already partitioned our data set into a training set and test set. This lets us judge whether a given model will generalize well to new data. However, using only two partitions may be insufficient when doing many rounds of hyperparameter tuning (which we don’t perform in this tutorial but it is always recommended to use a validation set).
Therefore, it is usually a good idea to create a so called validation set. Watch this short video from Google’s Machine Learning crash course to learn more about the value of a validation set.
We use k-fold crossvalidation to build a set of 5 validation folds with the function vfold_cv. We also use stratified sampling:
set.seed(100)
cv_folds <-
vfold_cv(train_data,
v = 5,
strata = price_category) We will come back to the validation set after we specified our models.